[BRANE Cut featured on RNA-Seq blog][Omic tools][bioRxiv preprint][PubMed/Biomed Central][BRANE Cut code][BRANE Omics]
Gene regulatory networks are somehow difficult to infer. This first work from an on-going work on BRANE Omics (termed BRANE *, for Biologically Related Apriori Netwok Enhancement) introduces an optimization method (based on Graph cuts, borrowed from computer vision/image processing) to infer graphs based on biologically-related a priori (including sparsity). It is succesfully tested on DREAM challenge data and an Escherichia coli network, with a specific work to derive optimization parameters from gene network cardinality properties. And it is quite fast.
Gene regulatory networks are somehow difficult to infer. This first work from an on-going work on BRANE Omics (termed BRANE *, for Biologically Related Apriori Netwok Enhancement) introduces an optimization method (based on Graph cuts, borrowed from computer vision/image processing) to infer graphs based on biologically-related a priori (including sparsity). It is succesfully tested on DREAM challenge data and an Escherichia coli network, with a specific work to derive optimization parameters from gene network cardinality properties. And it is quite fast.
BRANE Cut: Biologically-Related Apriori Network Enhancement with Graph cuts for Gene Regulatory Network inference (doi, BRANE Cut webpage, preprint)
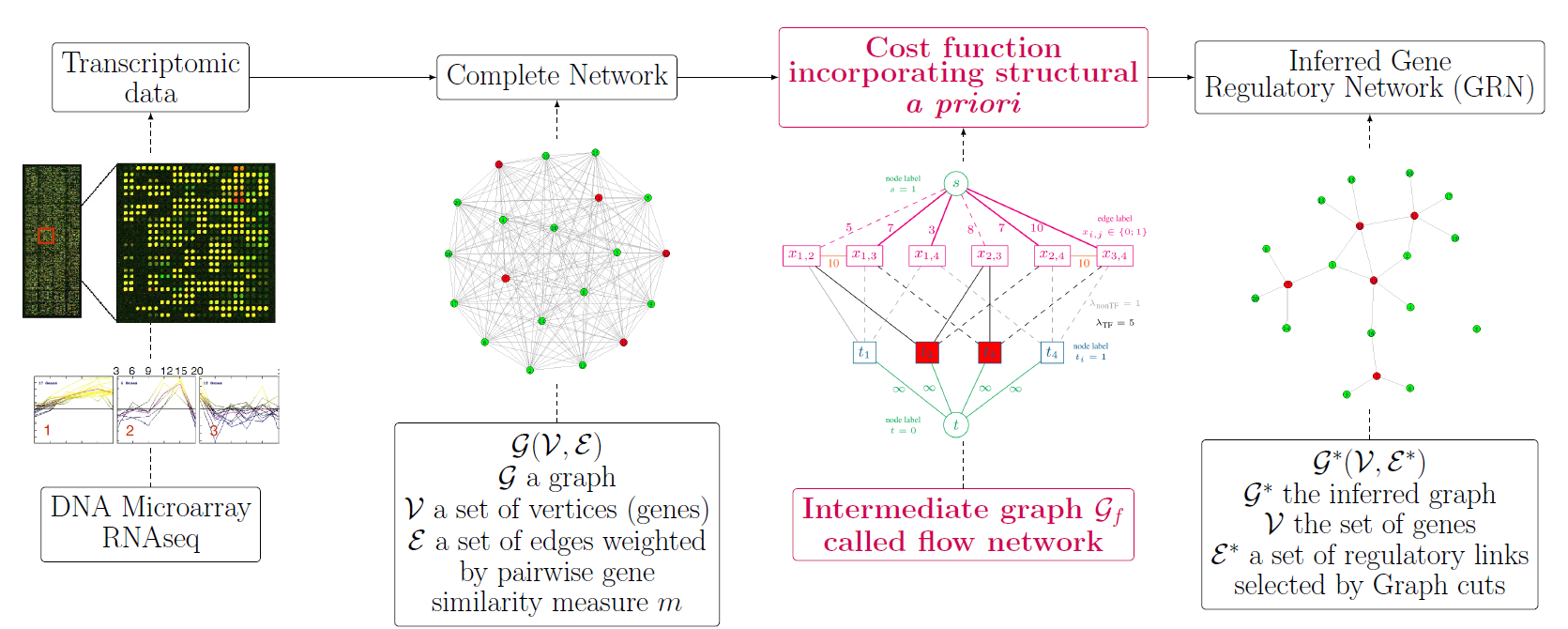
Background: Inferring gene networks from high-throughput data constitutes an important step in the discovery of relevant regulatory relationships in organism cells. Despite the large number of available Gene Regulatory Network inference methods, the problem remains challenging: the underdetermination in the space of possible solutions requires additional constraints that incorporate a priori information on gene interactions.
Methods: Weighting all possible pairwise gene relationships by a probability of edge presence, we formulate the regulatory network inference as a discrete variational problem on graphs. We enforce biologically plausible coupling between groups and types of genes by minimizing an edge labeling functional coding for a priori structures. The optimization is carried out with Graph cuts, an approach popular in image processing and computer vision. We compare the inferred regulatory networks to results achieved by the mutual-information-based Context Likelihood of Relatedness (CLR) method and by the state-of-the-art GENIE3, winner of the DREAM4 multifactorial challenge.
Results
Our BRANE Cut approach infers more accurately the five DREAM4 in silico networks (with improvements from 6 % to 11 %). On a real Escherichia coli compendium, an improvement of 11.8 % compared to CLR and 3 % compared to GENIE3 is obtained in terms of Area Under Precision-Recall curve. Up to 48 additional verified interactions are obtained over GENIE3 for a given precision. On this dataset involving 4345 genes, our method achieves a performance similar to that of GENIE3, while being more than seven times faster. The BRANE Cut code is available at: http://www-syscom.univ-mlv.fr/~pirayre/Codes-GRN-BRANE-cut.html.
Conclusions: BRANE Cut is a weighted graph thresholding method. Using biologically sound penalties and data-driven parameters, it improves three state-of-the art GRN inference methods. It is applicable as a generic network inference post-processing, due to its computational efficiency.
Keywords: Network inference, Reverse engineering, Discrete optimization, Graph cuts, Gene expression data, DREAM challenge.
Additional information of the BRANE Power page
Additional information of the BRANE Power page